Add Column (HASL) Dialog Box
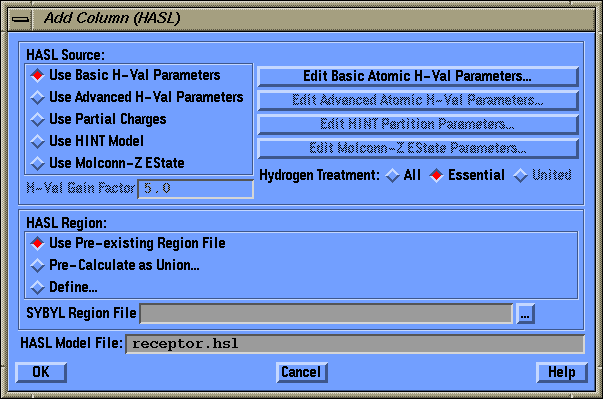
- HASL Source:
- radios: {Use Basic H-Val Parameters|Use Advanced H-Val Parameters|Use
Partial Charges|Use HINT Model|Use Molconn-Z EState}
- Designate the Data Source for the HASL model. The Basic H-Val parameter
set designates each atom type as having a parameter that is one of the
set {+1 0 -1}. The Advanced H-Val parameter set allows up to two parameters
for each atom type and will accept an integer value for either parameter.
Values between -5 and 5 are most reasonable. The current Partial Charge
set is also available as a model source. These will be multiplied by the
H-Val Gain Factor and rounded to the nearest integer. If you have a
HINT license,
you can use parameters from the HINT Model as the data source
for HASL or if you have a Molconn-Z
license you can use as input the Electrotopological State (EState). Again
these are multiplied by the H-Val Gain Factor and rounded to the nearest integer.
- H-Val Gain Factor
- field
- Enter a value for the "Gain" to be applied to HASL Models created using
"Partial Charges" or the "HINT Model". It is important that the "H-Val" parameter
used in creating the model be (more or less) in the range from -5 to 5. Thus,
the value of H-Val Gain Factor is critical. Reasonable values are on the order
of 2-5 for Partial Charge Models; 1-2 for HINT Models that do not include Solvent
Accessible Surface Area (i.e., Include SASA Factor in Parameter in the
HINT Parameters for HASL dialog box is "off"); 0.05-0.10
for HINT Models that include Solvent Accessible Surface Area; and 0.25-1.00
for EState Models.
- Edit Basic Atomic H-Val Parameters
- push button
- Use this command to examine and/or change the values of the H-Val
in the Basic parameter set. This button calls the Edit
Basic H-Val Parameters dialog box.
- Edit Advanced Atomic H-Val Parameters
- push button
- Use this command to examine and/or change the values of the H-Val
in the Advanced parameter set. This button calls the Edit
Advanced H-Val Parameters dialog box.
- Edit HINT Partition Parameters
- push button
- Use this command to examine and/or change the adjustable parameters used
by HINT to calculate the hydrophobic atom constant and atomic SASA sets for the
molecules. This button calls the HINT Parameters for
HASL dialog box.
- Edit Molconn-Z EState Parameters
- push button
- Use this command to examine and/or change the adjustable parameters used
by Molconn-Z to calculate the Electrotopological State (EState) atom constants for the
molecules. This button calls the Edit EState Parameters for
HASL dialog box.
- Hydrogen Treatment
- radios: {United|Essential|All}
- This menu describes how the HASL model will treat hydrogens;
there are three options: "United" (using an approach that treats
all hydrogens implicitly as part of their parent heavy atom),
"Essential" (using an approach that treats (only) polar
hydrogens explicitly), and "All". In general the
Essential option is best for many applications.
- HASL Region
- radio button set: {Use Pre-existing Region File|Pre-Calculate as Union...|Define...}
- Choose the method by which the SYBYL/HASL region is to be defined.
This region must be large enough to encompass the area of
interest in all molecules in the spreadsheet. Use
Pre-existing means that the region is defined by an
existing SYBYL region file on disk. Pre-Calculate
as Union... invokes the Calculate
SYBYL Region Automatically dialog box which defines
the region based on the union space spanned by the molecule
set. Define... invokes the Define
SYBYL Region dialog box which allows you to enter
the region as arbitrary boxes in cartesian space.
- SYBYL Region File
- field and push button
- This field specifies the name of the SYBYL Region File to
be used for creation of the HASL lattice. If the Region is
created by Pre-Calculate as Union... or Define... this
field will be filled by a return value from the associated
dialog boxes. If the Region is Use Pre-existing, you will
have to either enter the name of the region file or select
from the supplied list by using the ... button.
- HASL Model File
- field
- Identify the HASL model (.hsl) filename that you wish to use for this run.
The .hsl file maintains records of the associated parameters and filenames
for the model. As you are proceed in sequence from the creation of the
molecular HASL lattices through model creation and molecule prediction,
this parameter will be filled by default with this selection.

