HASL MCP Dialog Box
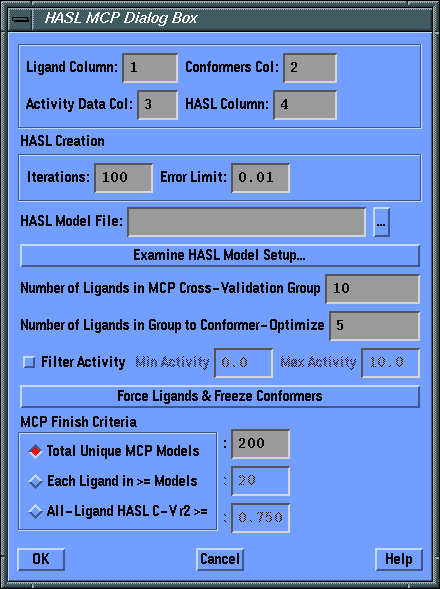
- Ligand Column
- field
- Enter the column number from the currently open table where the Ligand
Name Data for the molecule set can be found. This necessary column is
automatically created by the HASL MCP Database Dialog
command.
- Conformers Column
- field
- Enter the column number from the currently open table where the Conformer
Sequence Data for the molecule set can be found. This necessary column is
automatically created by the HASL MCP Database Dialog
command.
- Activity Data Column
- field
- Enter the column number from the currently open table where the Activity
Data for the molecule set can be found.
- HASL Column
- field
- Enter the column number from the currently open table where the molecule
lattice HASL results for the molecule set can be found.
- HASL Creation: Iterations
- field
- Enter the number of iterations to be performed in converging the HASL
model. The convergence will conclude when either the number of iterations
has been reached or model error is less than Error Limit.
- HASL Creation: Error Limit
- field
- Enter the minimum error threshold for the HASL model creation. The
convergence will conclude when either the number of iterations has been
reached or model error is less than Error Limit.
- HASL Model File
- field and push button
- Identify the HASL model (.hsl) file that you are using for this run.
The .hsl file maintains records of the associated parameters and filenames
for the model. If you are proceeding in sequence from the creation of the
molecular HASL lattices through model creation and molecule prediction,
this parameter will be filled by default with the name first chosen. Otherwise
you will have to enter it by name.
- Examine HASL Model Setup...
- push button
- If you wish to review the parameters used to create the set of molecular
HASL lattices, press this button. The resulting dialog, Examine
HASL Model Setup, lists the choices made in the Main HASL dialog.
- Number of Ligands in MCP Cross-Validation Group
- field
- The Multi-Conformer Protocol functions by randomly choosing subsets of ligands
in the spreadsheet (a model), and then optimizing these models through choices of the
conformers of each ligand. With this parameter the number of ligands to
include in each model is selected. Generally choose a number between 5 and 10.
- Number of Ligands in Group to Conformer-Optimize
- field
- As the models are refined, the MCP systematically optimizes ligands by
trying all conformers of that ligand. The MCP starts with the poorest predicted
ligand (from a cross-validation of the model set), followed by the second
poorest predicted ligand, and so forth. This parameter sets the number of
ligands in each model that will be so optimized. The assumption is that
the best few predicted ligands may not benefit from conformer-optimization as the
initial model selection probably randomly selected one of the better conformers.
- Filter Activity
- check box
- If this box is checked "on" the activity range identified by
Min Activity and Max Activity will be used to filter the Molecular Spreadsheet,
i.e., ligands whose activity range falls outside of these values will not
be included in the ligand pool or in any models.
- Min Activity
- field
- Minimum activity for filter.
- Max Activity
- field
- Maximum activity for filter.
- Force Ligands & Freeze Conformers...
- push button
- If you wish to "force" certain ligands to be in each model, or
"freeze" the conformations of those ligands, e.g., if a crystal conformation
is known, then select this option to activate the MCP
Force Ligands & Freeze Conformers Dialog.
- MCP Finish Criteria
- radios and fields: {Total Unique MCP Models|Each Ligand in >= Models|All-Ligand HASL C-V r2>=}
- This collection of parameters offers three ways for the MCP calculation
to exit "gracefully". Note, however, that the results of each completed
model (and sub-model) are written to a results file, such that there is
no penalty for ending the MCP prematurely with a "control-C" in the Sybyl text window
if you feel that the ligand/conformation space has been adequately sampled.
- Total Unique MCP Models: the MCP will end when this number of
models has been calculated.
- Each Ligand in >= Models: the MCP will end when each ligand in the
ligand pool has been examined at least this number of times in unique models.
- All-Ligand HASL C-V r2>=: the MCP will end when the cross-validated
r2 for an All-Ligand HASL model exceeds this value. An all-ligand
HASL validation is run each time the ligand count criteria (above) is incremented, i.e.,
when each ligand has appeared in at least one model, when each ligand has appeared
in at least two models, etc.

